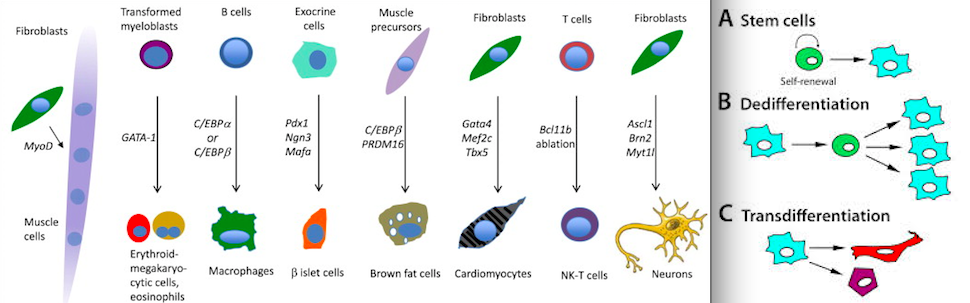
Epigenetic manipulation refers to changing genetic expression of certain genes(in this case height increasing genes) via nutritional or mechanical means. This manipulation can occur by altering histones, chromatin folding, methylation, telomere length, etc.
The paper below indicates that altering epigenetics can powerfully influence but the question is how to determine the mechanical and nutritional methods that can influence these genes.
Epigenetic heredity of human height
“Genome‐wide SNP analyses have identified genomic variants associated with adult human height. However, these only explain a fraction of human height variation, suggesting that significant information might have been systematically missed by SNP sequencing analysis. A candidate for such non‐SNP‐linked information is DNA methylation. Regulation by DNA methylation requires the presence of CpG islands in the promoter region of candidate genes{So any height increase genes that have CpG islands can be altered by DNA methylation}. Seventy two of 87 (82.8%), height‐associated genes were indeed found to contain CpG islands upstream of the transcription start site, which were shown to correlate with gene regulation. Consistent with this, DNA hypermethylation modules{hypermethylation can result in transcription silencing which can be inherited by daughter cells(a daughter is the cell formed by mitosis)-mitosis occurs in the growth plate most heavily in the proliferative zone} were detected in 42 height‐associated genes, versus 1.5% of control genes, as were dynamic methylation changes and gene imprinting. Epigenetic heredity thus appears to be a determinant of adult human height. Modulation of DNA methylation are candidate to mediate environmental influence on epigenetic traits. This may help to explain progressive height changes over multiple generations, through trans‐generational heredity of progressive DNA methylation patterns.”
Some height increase genes identified in multiple studies:
(ACAN, BCAS3 also known as TBX2, EFEMP1, HHIP, HMGA1, HMGA2, LCORL, NCAPG, PLAGL1, PTCH1, SOCS2, SPAG1, UQCC also known as GDF5, ZBTB38, ZNF678)
“Genes close to the SNP most strongly associated with body size were shown to encode extracellular matrix components, proteases, cell cycle controllers, transcription factors and signaling molecules”
Table 1 in the paper gives a list of height related genes. Here’s a list of genes related to height increase and whether you want to upregulate or downregulate the genes relative to height increase.
“Functionally‐relevant DNA methylation patterns were thus candidates to be associated with adult stature subgroups in addition to DNA sequence variants. Functionally‐relevant DNA methylation patterns may affect selective mechanisms, thus behaving as true hereditary traits. Consistent with this, a metastable epigenetic heredity of the DWARF1 locus was shown to affect plant size and this phenotype was inherited through mitosis and meiosis”
“DNA methylation patterns can keep record of the nutritional status and affect, in turn, morphometric parameters. Modifications of DNA methylation patterns in growth‐related genes can be inherited trans‐generationally, through incomplete erasure of epigenetic patterning in the germline.”
“Genomic imprinting defects are associated with developmental disorders, including Silver‐Russell, Beckwith‐Wiedemann, and Prader‐Willi syndromes. Genomic imprints are affected by environmental factors, and also associate with several human cancers.”
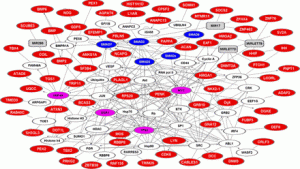
“Proteins are represented as nodes (hubs), the biological relationships between the nodes (edges) are represented as lines. Height‐associated proteins are in red; linker proteins are in white; miRNA are in gray. Major hubs are in magenta; SMAD isoforms are in blue.”
“72 of 87 height‐associated genes (82.8%) were found to contain at least one CpG island in the 2,000 bp upstream of the transcription start site (TSS) (99 CpG islands overall) . Notably, in all CpG islands‐associated height genes, CpG islands overlapped with the TSS, supporting an actual regulatory role in gene transcription”
Notable genes regulated by DNA Methylation according to Table 2 include: BMP2, BMP6, and SOCS2 as well as several not normally associated with height increase. Notable genes regulating DNA Methylation(that is the control the methylation status of height related genes) include: DNMT3A, DOT1L, HMGA1, HMGA2.
“five genes (ACAN, ANKS1, FBP2, NACA2, ZBTB38) were found to have no evidence of DNA methylation. The remaining genes (94.3%) were shown to undergo broad changes of DNA methylation levels across experimental conditions”
“CpG island methylation in the BMP2 promoter causes loss of BMP‐2 protein expression in transformed cells{You would NOT want this if your desire was to have your child grow taller}. Shut‐down of the BMP6 gene by promoter methylation was observed in malignant lymphomas”
“c‐Myc regulates at least seven height‐associated genes (CDK6, COIL, HMGA1, LIN28B, RBBP8, RPS20, TRIM25/EFP), and its binding to genomic loci is dependent on chromatin structure and CpG methylation.”
“The Beckwith‐Wiedemann syndrome is caused by deregulation of imprinted genes within the 11p15 chromosomal region, i.e., KIP2, H19 and LIT1, whether alone or as interacting regulatory units . Hypermethylation at the 11p15 telomeric imprinting control region (ICR1), are observed in about 5 to 10% of affected patients. Both H19 and LIT1, which encode untranslated RNAs, and IGF2 are either maternally imprinted genes with growth enhancing activity or paternally imprinted genes with growth suppressing activity.”
“Affected children reach an average height of 2.5 SD above the mean at or after puberty, and their growth velocity is above the ninetieth percentile until 4–6 years of age.”
“Up to 60% of cases of Silver‐Russell [dwarfism] syndrome are caused by hypomethylation at the ICR1 on chromosome 11p15, involving the H19and IGF2 genes”<-so underexpression of ICR1 is good for height and overexpression of ICR1 is bad for height if hypermethylation transcriptionally silences expression and hypomethylation increases it.
“c‐Myc regulates the cell cycle, and plays a major role in cell growth during interphase, by regulating genes required for the production of energy and metabolites. The c‐Myc network widely interacts with those driven by other major hubs. c‐Myc is repressed by transforming growth factor β (TGF‐β) through the binding of SMAD3 to the MYC promoter. p53 represses c‐Myc through the induction of the tumor suppressor miR‐145. c‐Myc amply interacts also with the ER network: almost all of the acutely estrogen‐regulated genes with roles in cell growth are c‐Myc targets. Notably, estrogen‐mediated activation of rRNA and protein synthesis depends on c‐Myc. Equally c‐Myc dependent is the estrogen‐induced suppression of apoptosis caused by growth factor deprivation”
“p53 regulates the expression of target genes that modulate chromatin structure and function, cell growth, aging and apoptosis. p53 interacts with components of multiple different histone remodeling complexes, including CBP/EP300 (CBP/p300), GCN5, PCAF, and SETD7 modifying histones at the promoters. p53 also controls DNA methylation levels, and that this affects genome stability”
“ERα regulates at least eight height‐associated genes (BCAS3, BMP2, BMP6, DCC, GLT25D2, PENK, RBBP8, TRIM25/EFP).”
“ERα blockade diminishes the secretion of endogenous growth hormone, the key hormone regulator of linear growth in childhood. This action is mediated by SOCS‐2. The ERα network widely interconnects with the p53, Hh and BMP/TGF‐β pathways. p53 regulates ER expression through transcriptional control of the ER promoter”